//
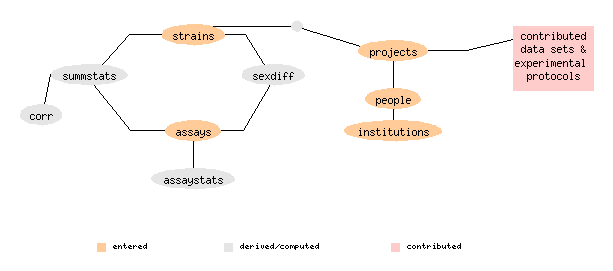
// A simple database ER diagram using proc annotate.
// Entity bubbles support mouseover and clickthru, when HTML includes
// client-side image map (see er.csmap)
//
// pl -gif er.htm -o er.gif -csmap -mapfile er.csmap -scale 0.8
#set URL = "http://aretha.jax.org/pub-cgi/phenome/mpdcgi?rtn=download/tabledetails"
#procdef annotate
#saveas A
backcolor: lightorange
ellipse: yes
// outline: yes
arrowheadsize:
// 3rd arrow - strains to projects (must be done before for clean line)..
#proc annotate
location: 3 3
arrowhead: 5.5 2.8
#proc annotate
#clone A
location: 3 3
arrowhead: 1.7 2.5
arrowhead2: 4 2.5
clickmapurl: @URL&table=strains
clickmaplabel: mouse strains having data in the MPD
text: strains
// 3rd arrow - assays to assaystats (must be done before for clean line)..
#proc annotate
location: 3 1.8
arrowhead: 3 1.2
#proc annotate
#clone A
location: 3 1.8
arrowhead: 1.7 2.5
arrowhead2: 4 2.5
clickmapurl: @URL&table=assays
clickmaplabel: description of each MPD measurement
text: assays
#proc annotate
#clone A
location 1.7 2.5
arrowhead: 1.1 2
backcolor: gray(0.9)
clickmapurl: @URL&table=summstats
clickmaplabel: summary statistics computed for each measurement / strain / sex
text: summstats
#proc annotate
#clone A
location 1.1 2
backcolor: gray(0.9)
clickmapurl: @URL&table=corrstrong
clickmaplabel: correlations computed between all possible pairs of MPD measurements
text: corr
#proc annotate
#clone A
location 4 2.5
backcolor: gray(0.9)
clickmapurl: @URL&table=sexdiff
clickmaplabel: sex differences computed for each measurement / strain
text: sexdiff
#proc annotate
#clone A
location: 3 1.2
backcolor: gray(0.9)
clickmapurl: @URL&table=assaystats
clickmaplabel: summary statistics computed for each measurement
text: assaystats
#proc annotate
#clone A
location: 5.5 2.8
arrowhead: 5.5 2.2
arrowhead2: 7 3
clickmapurl: @URL&table=projects
clickmaplabel: phenotyping and genotyping projects
text: projects
#proc annotate
#clone A
location: 5.5 2.2
arrowhead: 5.5 1.8
clickmapurl: @URL&table=people
clickmaplabel: participating investigators
text: people
#proc annotate
#clone A
location: 5.5 1.8
clickmapurl: @URL&table=institutions
clickmaplabel: institutions associated with investigators
text: institutions
#proc annotate
#clone A
location: 7.5 3
ellipse: no
backcolor: pink
clickmapurl: @URL&table=contributed-data
text: contributed
data sets &
experimental
protocols
// projstrains (dot)
#proc annotate
#clone A
backcolor: gray(0.9)
backdim: 4.3 3.173 0.1 0.1
clickmapurl: @URL&table=projstrains
clickmaplabel: many-to-many mapping between projects and strains
////////////////////////////
#proc legendentry
sampletype: color
details: lightorange
label: entered
#proc legendentry
sampletype: color
details: gray(0.9)
label: derived/computed
#proc legendentry
sampletype: color
details: pink
label: contributed
#proc legend:
location: 2 0.5
format: singleline
