How to download and try this example
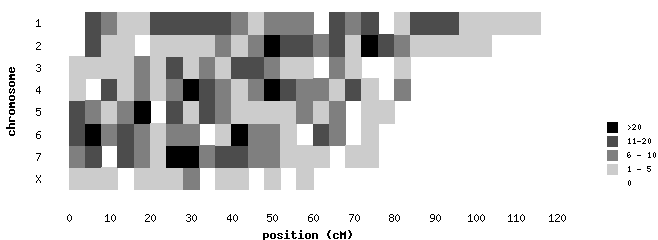
Usage: pl -gif [-map] heatmap2.htm This color grid involves processing raw data by counting occurances within ranges, then mapping counts to colors by range. Uses data file snpmap.dat . For brevity, only chromosomes 1-7 and X are represented in this example.
#set SYM = "radius=0.08 shape=square style=filled" #setifnotgiven CGI = "http://ploticus.sourceforge.net/cgi-bin/showcgiargs" // read in the SNP map data file.. #proc getdata file: snpmap.dat fieldnameheader: yes // group into bins 4 cM wide.. filter: ##set A = $numgroup( @@2, 4, mid ) @@1 @@A // set up the plotting area #proc areadef rectangle: 1 1 6 3 areacolor: white yscaletype: categories clickmapurl: @CGI?chrom=@@YVAL&cM=@@XVAL ycategories: 1 2 3 4 5 6 7 X yaxis.stubs: usecategories // yaxis.stubdetails: adjust=0.2,0 //yaxis.stubslide: 0.08 yaxis.label: chromosome yaxis.axisline: no yaxis.tics: no yaxis.clickmap: xygrid xrange: -3 120 xaxis.label: position (cM) xaxis.axisline: no xaxis.tics: no xaxis.clickmap: xygrid xaxis.stubs: inc 10 xaxis.stubrange: 0 // xaxis.stubdetails: adjust=0,0.15 // set up legend for color gradients.. #proc legendentry sampletype: color details: gray(0) label: >20 tag: 21 #proc legendentry sampletype: color details: gray(0.3) label: 11-20 tag: 11 #proc legendentry sampletype: color details: gray(0.5) label: 6 - 10 tag: 6 #proc legendentry sampletype: color details: gray(0.8) label: 1 - 5 tag: 1 #proc legendentry sampletype: color details: gray(1) label: 0 tag: 0 // use proc scatterplot to count # of instances and pick appropriate color from legend.. #proc scatterplot yfield: chr xfield: cM cluster: yes dupsleg: yes rectangle: 4 1 // display legend.. #proc legend location: max+0.7 min+0.8 textdetails: size=6