How to download and try this example
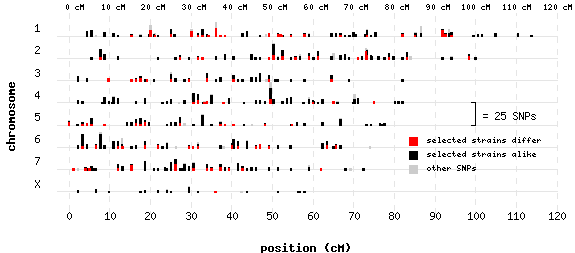
Usage: pl -gif snpmap1.htm Uses data file snpmap.dat . For brevity, only chromosomes 1-7 and X are represented in this example.
// set demo defaults.. #setifnotgiven SLIST = "@9,@11" #setifnotgiven CGI = "http://ploticus.sourceforge.net/cgi-bin/showcgiargs" // SLIST is a comma-delimited list of field numbers (eg @4) indicating strain(s) of interest // (the integers correspond to field positions in the SNP map data file). // read in the SNP map data file.. #proc getdata file: snpmap.dat fieldnameheader: yes #if @SLIST != "" // if we are concentrating on strain(s) of interest, run some processing to flag // entries where strain(s) of interest are all present, and to flag entries where // strains of interest have different alleles.. // showresults: yes filter: // see if alleles of interest are all present, and if there are differences ##set LIST = $expand( "@SLIST" ) ##set ALLPRESENT = 0 ##set DIFF = 0 ##if = !in @@LIST ##set ALLPRESENT = 1 ##endif ##if $homogeneous( @@LIST ) = 0 ##set DIFF = 1 ##endif @@1 @@2 @@ALLPRESENT @@DIFF #endif // set up the plotting area #proc areadef rectangle: 1 1 6 3 yscaletype: categories clickmapurl: @CGI?chrom=@@YVAL&cM=@@XVAL ycategories: 1 2 3 4 5 6 7 X yaxis.stubs: usecategories yaxis.stubslide: 0.08 // yaxis.stubreverse: yes yaxis.label: chromosome yaxis.axisline: no yaxis.tics: no yaxis.grid: color=gray(0.9) yaxis.clickmap: xygrid xrange: -3 120 xaxis.label: position (cM) xaxis.grid: color=gray(0.9) width=0.4 style=1 dashscale=3 xaxis.axisline: no xaxis.tics: no xaxis.clickmap: xygrid xaxis.stubs: inc 10 xaxis.stubrange: 0 xaxis.stubdetails: adjust=0,0.15 // do 2nd x axis across top #proc xaxis location: max axisline: no tics: no stubs: inc 10 stubdetails: adjust=0,0.2 size=6 stubformat: %3.0f cM stubrange: 0 #if @SLIST = "" #set ALLCOLOR = teal #else #set ALLCOLOR = gray(0.8) #endif // use proc scatterplot to cluster duplicates upward, effectively creating little bars.. #proc scatterplot yfield: chr xfield: cM symbol: shape=square radius=0.01 fillcolor=@ALLCOLOR style=filled cluster: yes clustermethod: upward clusterfact: 0.4 #saveas A #if @SLIST != "" // overlay little black bars counting entries where all strains of interest are present.. #proc scatterplot #clone A select: @@3 = 1 symbol: shape=square radius=0.01 fillcolor=black style=filled // overlay little red bars counting entries where strains of interest have different alleles.. #proc scatterplot #clone A select: @@3 = 1 && @@4 = 1 symbol: shape=square radius=0.01 fillcolor=red style=filled #endif // do legend if appropriate.. #if @SLIST != "" #set N = $count( "*", @SLIST ) #if @N = 1 #proc legendentry sampletype: color details: black label: involving strain(s) #proc legendentry sampletype: color details: @ALLCOLOR label: not involving strain(s) #elseif @N > 1 #proc legendentry sampletype: color details: red label: selected strains differ #proc legendentry sampletype: color details: black label: selected strains alike #proc legendentry sampletype: color details: @ALLCOLOR label: other SNPs #endif #proc legend location: max-1.3 min+0.8 textdetails: size=6 swatchsize: 0.075 #endif // draw scale bracket.. #proc line linedetails: width=0.4 points: 100(s) 5(s) 100(s) 4(s) 100(s) 5(s) 99(s) 5(s) 100(s) 4(s) 99(s) 4(s) #proc annotate location: 102(s) 5(s) textdetails: align=L size=7 adjust=0,0.05 text: = 25 SNPs